Fast Fat/Water Decomposition Using GPU Computation and Newton's Method
David Johnson, Sreenath Narayan, Chris Flask, David Wilson
ISMRM 2010, Poster Presentation #3156. The poster is available here [PDF].
Abstract
An improved fat/water estimation technique was developed using a formulation similar to the Iterative Decomposition of Water and Fat with Echo Asymmetry and Least-squares estimation method and Graphics Computational Units (IDEAL-GPU). The IDEAL-GPU technique produced robust fat and water images quickly and efficiently using a vectorized equation implemented on graphics cards. In addition, our implementation used binary weighted planar extrapolation for robust estimation in the face of large field variations on a high field, small animal scanner. Fast computation will become even more significant as the trend towards high resolution, whole body mouse and human scanning continues.
Introduction
We are developing quantitative MRI techniques to quantify fat depots (e.g., visceral, subcutaneous, hepatic, muscular) to determine the role of genetic, environmental, and therapeutic factors on lipid accumulation, metabolism, and disease states. We have shown previously that conventional clinical scanners can provide sufficient image quality for rats and larger animals [1]. However, high field MRI scanners (7T-11T) are needed to produce the high resolution images that provide the basis for accurate delineation between visceral and subcutaneous lipid compartments in mice [2]. However, the data processing time is significant because 3-6 image sets at variable echo times must be acquired resulting in >1GB of data. This requires over 1 hour of processing time for each animal. The purpose of this study was to develop a method to more quickly produce fat and water estimates enabling rapid MRI phenotyping. Our initial results show a 2- to 12-fold reduction in processing time when GPU computations are used, which greatly eases the burden of the IDEAL reconstruction time.
Methods
Three 26 week old C57BL/6J male mice were scanned using an asymmetric spin echo acquisition (aSE) for IDEAL [3], which was implemented for imaging mice on a 7T/30cm Bruker Biospec scanner [4]. We reformulated the IDEAL equations using vectorization and implemented the algorithm using a Matlab GPU library (Accelereyes Jacket). In contrast to previous IDEAL per-pixel algorithms, vectorized IDEAL evaluates the residuals (J) of each estimate of the field inhomogeneity, ψ(x), in each of the pixels of the image (x, x+1, x+2, ... x+N*M), all at once. The observation matrix A ([1 e^(j2π*TE(1)*1050Hz); 1 e^(j2π*TE(2)*1050Hz); 1 e^(j2π*TE(3)*1050Hz)]), and a temporary matrix T (the per-element product of the observed signals S(x,TE) and ψ(x)), are also calculated, as per Eqs (1) and (2).
(1)
(2)
The first and second partial derivatives of J with respect to ψ were evaluated analytically, and the residuals were independently minimized in each pixel using our GPU implementation of Newton's method. Newton's method "jumps" to the minimum (apex) of a parabola determined by the ratio of the first and second derivatives (Fig 1). The method was tested on the three mouse datasets each with TEs corresponding to p/6, 5p/6, and 3p/2 radian shifts between fat and water. IDEAL-GPU was run 11 times and the execution times excluding the first were averaged. The effects of different data sizes on the CPU and GPU execution times were tested by downsampling the images from the acquired 512x512 size along the phase encoding direction.
Results
The execution time of running the reconstruction was reduced by 2- to 12-fold using our GPU algorithm, corresponding to a reduction from 4 minutes per image to <10 seconds per image (Fig 2). The relative speedup of the GPU increased to almost 12X for larger images (Fig 3), but no additional gains were observed for images bigger than 512x512 pixels. The use of single precision floating point calculations on both CPU and GPU did not visibly affect the quality of the reconstructed images.
Discussion
IDEAL provides excellent fat and water estimates, but the processing thrice the data of one acquisition is a burden. Using 6 or more TEs for multi-peak IDEAL [5] is an even greater computational challenge. IDEAL-GPU addresses these problems by reformulating the estimation as a series of efficient matrix multiplications. IDEAL-GPU is not dependent on a specific video card, and we anticipate further speedups with the development of newer video cards with higher clock rates and more processor elements. Also, additional video cards can be used in parallel to reconstruct multiple images simultaneously, allowing for even more scalability.
References and Acknowledgements.
[1] Johnson et al. JMRI 2008; 28(4):915-927. [2] Johnson et al. accepted for publication JMRI 2009. [3] Reeder et al. MRM 2005; 54(3): 636-644. [4] Johnson et al., ISMRM 2009:2682. [5] Kijowski et al. JMRI 2008; 29(2): 436-442. This work was supported by NCI R24-CA110943 (Northeast Ohio Animal Imaging Resource Center), NIH F30DK082132, NIH R01-EB004070, and NIH 1T32EB007509-01 (Interdisciplinary Biomedical Imaging Training Program).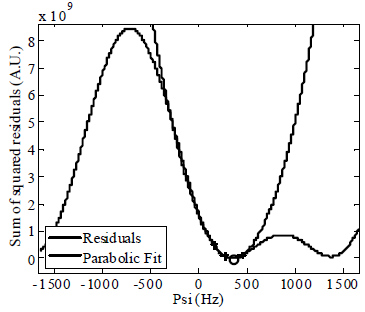
Fig 1. Newton's method finds the minimum quickly by fitting a local parabola.
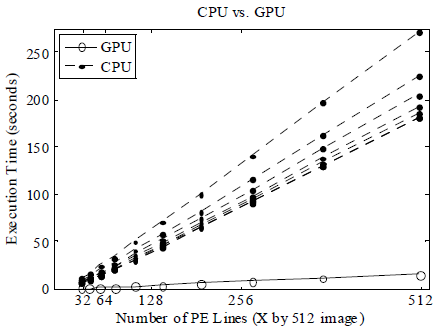
Fig 2. GPU execution times (solid line) were much faster than the CPU, even when multiple CPUs were used (multiple broken lines).
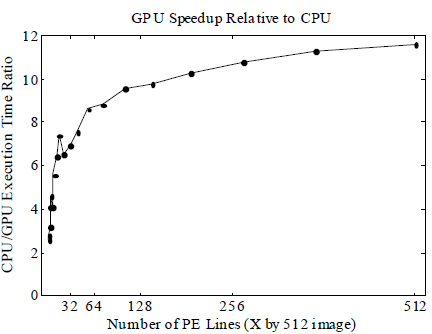
Fig 3. The GPU advantage over the CPU continued to improve for up to 512x512 images.